Table
of Contents
The
Main Panel
The
main panel
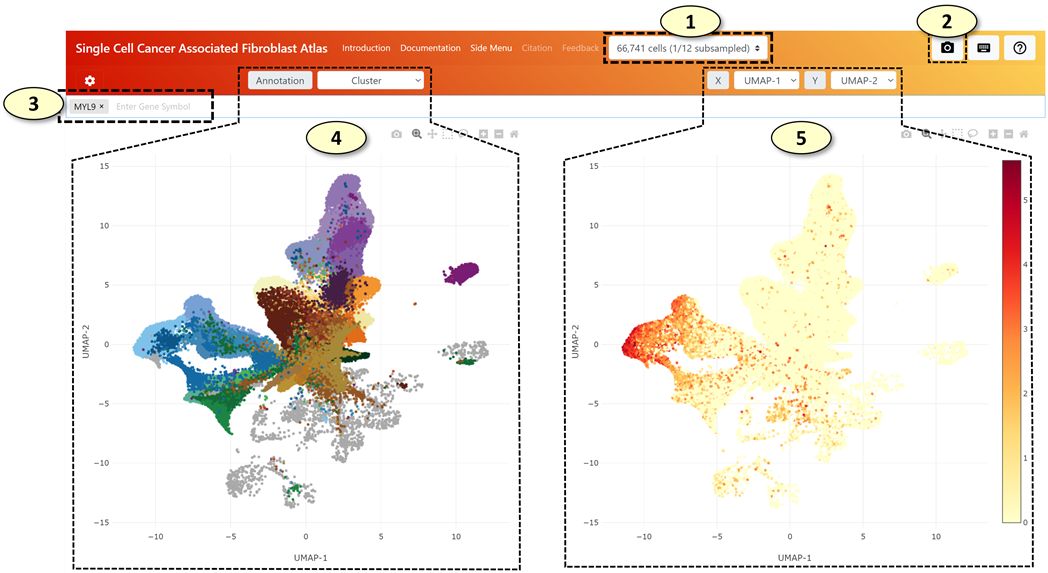
1.
[Select
a Subsampling Level] Select
how many cells will be loaded on the web application.
l Loading all cells (838,388
cells) from the dataset will require 1.7 GB of memory (a peak memory usage of
3.8 GB). Therefore, we recommend loading subsampled datasets on mobile
browsers, while loading the full dataset only on the desktop.
2.
[Capture]
Capture and
save the current view as a PNG image file
3.
[Gene
Search Box] Users
can explore single-cell gene expression patterns of each gene by typing a valid
gene symbol in this gene search box.
1)
The
search box has an auto-completion feature that suggests a list of partially
matched gene symbols to the user as a gene symbol is typed.
2)
The
summarized gene expression pattern of a gene is instantly visualized as it is
being typed to aid more efficient exploration of our dataset without
downloading additional data from the server.
3)
Once
an enter key is pressed, a tag representing the gene symbol (a gene symbol tag)
is added to the gene search box. Next, a compressed data chunk containing an
array of gene expression values of the gene symbol for the current subsampled
(or all) cells is downloaded from our database, parsed in the user's web
browser, and displayed on the web application.
4)
Up
to 200 genes can be simultaneously loaded in the web
application by adding gene symbol tags in the gene search box one by one. A
summarized gene expression pattern of each gene can be visualized by moving a
mouse pointer over each gene symbol tag without downloading any additional data
from our web database.
5)
By
clicking the 'X' symbol left to the gene symbol in each gene symbol tag, a gene
symbol tag can be removed from the main gene search box. The removal of the
gene symbol tag will lead to the removal of the single-cell-level gene
expression values for the gene symbol from the memory, thus freeing unnecessary
memory used by the web application
4.
[UMAP
Plot for Cell Annotation]
Users can explore annotation labels of the cells on a UMAP graph. The type of
annotation labels can be changed by selecting a different annotation in the
selection tab located above the UMAP plot.
5.
[UMAP
Plot for Gene Expression] This
UMAP graph will visualize normalized and log-transformed gene expression values
of individual single cells using the UMAP coordinates of each cell.
l Clicking a cell on either of
the UMAP graphs will trigger downloading of the list of cell marker genes for
the cluster to which the cell belongs, which will be subsequently visualized in
the [Cell Marker Panel]. The list of cell markers can be downloaded as a
CSV file by clicking the 'Download' button located above the table displaying
the marker gene list.

l Users can zoom in into a specific
region of the UMAP plot by selecting the region when a magnifying glass symbol
is active on the toolbar on the right upper side of the UMAP graph.
l The two UMAP plots, the cell
annotation, and gene expression UMAP plots will always display the same region
of a graph, visualizing cell annotation labels and gene expression levels of
the cells, respectively. The change of view of one UMAP plot will be
automatically applied to that of the other UMAP plot.
l Additionally, users can
change x-axis or y-axis values from UMAP coordinates to gene expression values
of a gene of interest by (1) selecting the 'gene' option in the selection tab
next to either the 'X' or 'Y' labels above the [UMAP Plot for Gene
Expression] plot and (2) clicking the gene symbol tag in the gene search
box. After the change of an axis, the axis label will be updated from 'UMAP-1'
(or 'UMAP-2') to the gene symbol that has been selected to represent the axis. The
axis can be changed back to the UMAP coordinates by selecting the 'UMAP-1' (or
'UMAP-2') option in the selection tab.